- Home
- About the Department
- Staff and contacts
- Study
- Research
- Research Groups
- Laboratory of experimental hematology and stem cells
- Laboratory of Interaction of Cells with Nanomaterials
- Lymphoma Lab
- The Laboratory of Molecular Mechanisms in Blood Diseases
- IronLab - Laboratory of Iron Metabolism
- MicroRNA and transcription factors in regulation of hematopoesis and leukemogenesis
- Laboratory of Proteomics and Extracellular Vesicles
- Publications
- Events
- Research Groups
- Postgraduate study
- Links
MicroRNA and transcription factors in regulation of hematopoesis and leukemogenesis - Copy
Mentor: Mgr. Vít Pospíšil, Ph.D.

Email: vitek_pos@hotmail.com
Mob: (+420) 732 953372
Research associate/postdoc:
Pavle Krsmanovic, Msc, PhD

Email: krsmanovic.pavle@gmail.com
Graduate student: Kamila
Chramostová, Bc.

Email:
kamila.chramostova@natur.cuni.cz
Alumini: Jan Valečka, Msc., Research associate
Current affiliation: CIML Centre d'Immunologie de Marseille-Luminy, France

Email: hojaval@gmail.com
Mob: (+420) 224 965863
We are lab union with Stopka lab, http://stopka-lab.lf1.cuni.cz/ , shearing knowledge, equipment and some projects.
Projects:
1. Role of hematopoietic master transcription factor PU.1 upon blood cell differentiation
We and others demonstrate that distinct levels of
PU.1 navigate differentiation of hematopoietic stem cells and
progenitors into different blood cell lineages. Different levels of PU.1
activate different lineage specific genes and microRNAs. However, the
molecular mechanisms of activation and repression of different genes
mediated by different levels of single transcription factor have not
been described yet and is challenging biological question. The
approaches as functional testing of
putative enhancers by reporter constructs, their 3D
interaction, role of chromatin modification, gene
manipulations etc. are currently being employed.
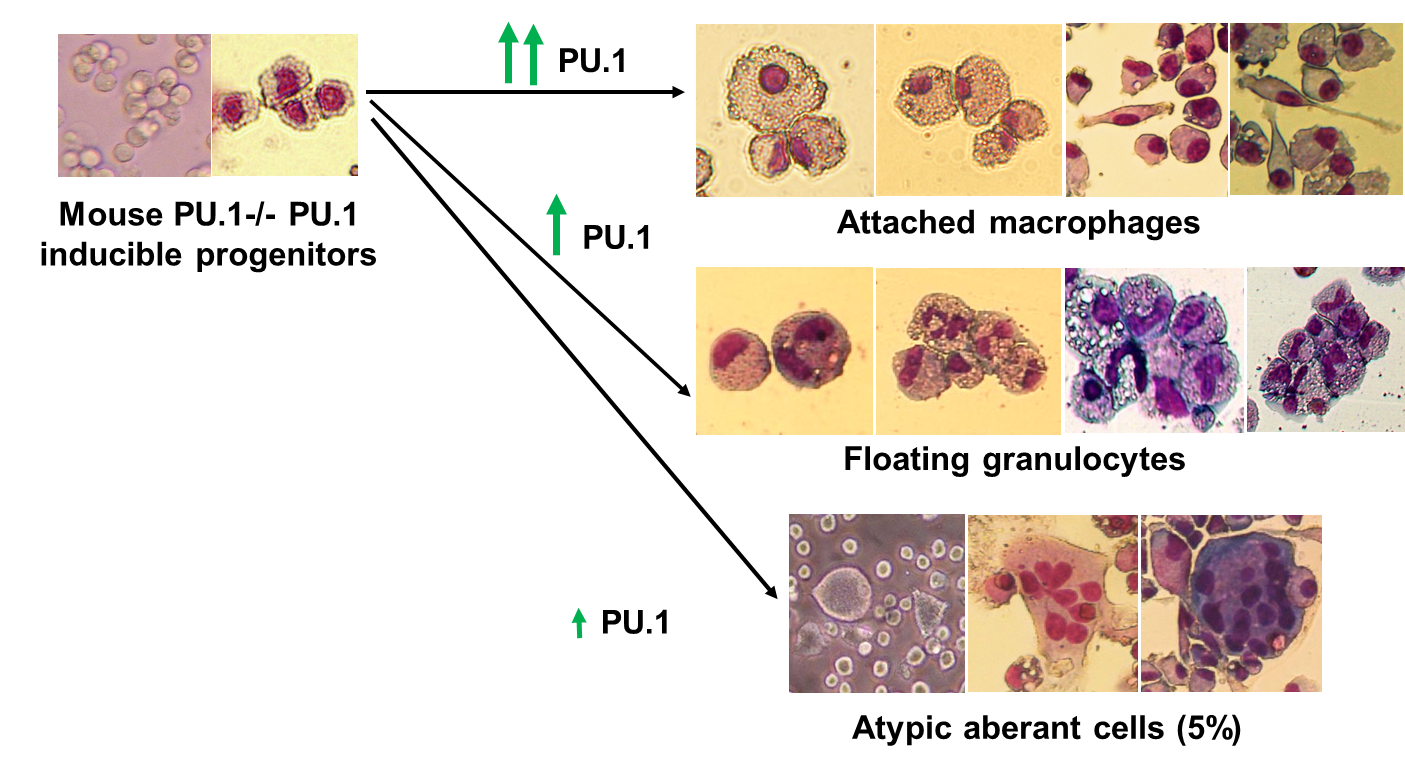
2. Mutual regulation between microRNAs and transcription factors upon blood cell differentiation
Discovery of microRNAs brought missing feedback
motive to pure transcription factors gene regulatory network. However,
transcription factors, microRNAs, or their target genes don’t act
independently but form multilevel mutual gene regulatory networks, where
change in expression of one gene can affect dozens of other genes or
vice versa one gene can be functionally substituted by others. We
described such mutual TF - microRNA regulatory motive upon macrophage
differentiation of myeloid progenitors and Acute Myeloid leukemia
patients blasts (see schema). Question is
whether such regulatory circuit is cell type specific, functional in
other blood cell lineages or affected in other types of leukemia or
lymphoma.
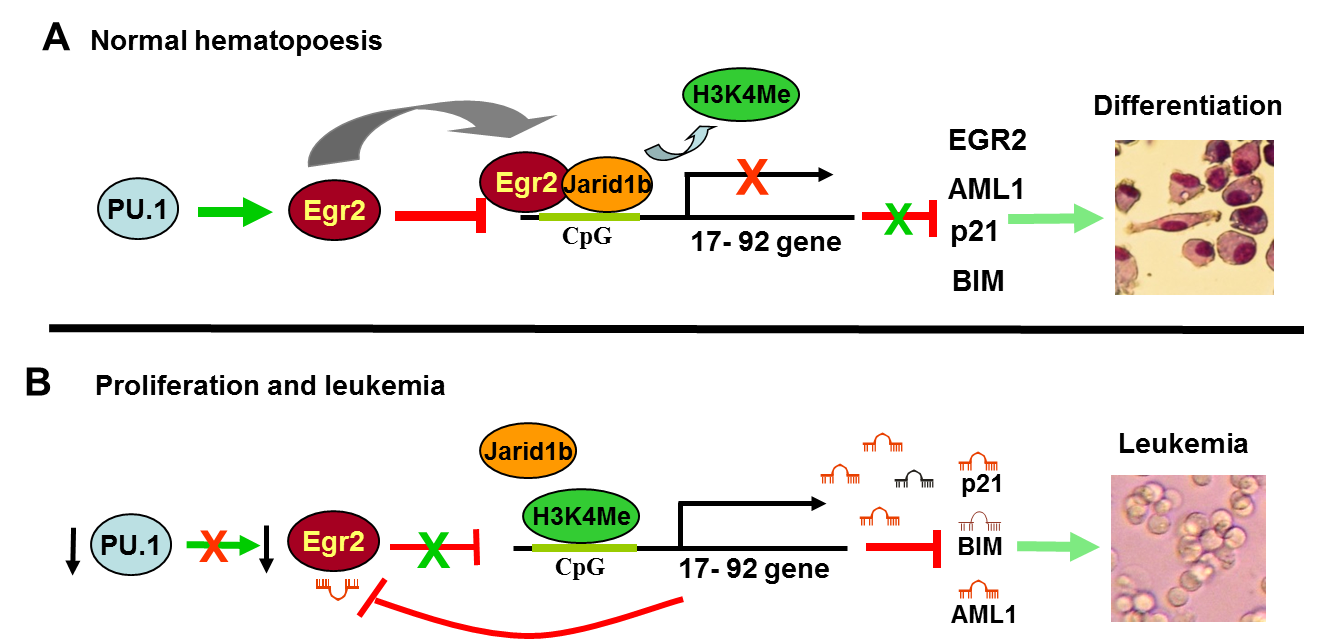
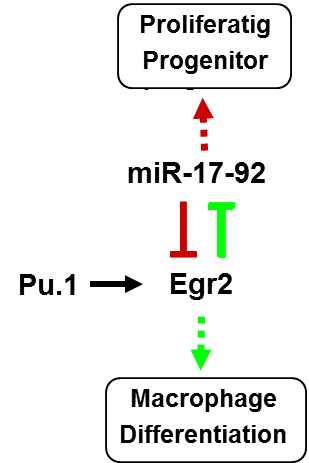
Adapted from Pospisil et al., EMBO journal, 2011
3. The development of sensitive methods for early detection of lymphomas of the central nervous system by oncogenic microRNA in CSF and plasma
Lymphoma involving CNS represents one of the most aggressive malignancies with poor prognosis and high mortality. Their current diagnosis suffers from both low sensitivity and specificity and is only able to detect well-established lymphoma. The prediction, early detection, response evaluation and monitoring of CNS lymphoma is urgently needed.
Our preliminary data indicate that we identified specific set of oncogenic microRNAs that can several months in advance identify CNS lymphoma involvement with high specificity, reflects therapy efficiency or predicts progression or relapse, thus substituting the weakness of current diagnostic methods and prognostic tools. The diagnostic method is further developed and tested.
This project is in cooperation with General university hospital in Prague and University hospital Kralovske Vinohrady, represented by Prof. Marek Trneny PhD, and MUDr. Heid Mocikova, PhD
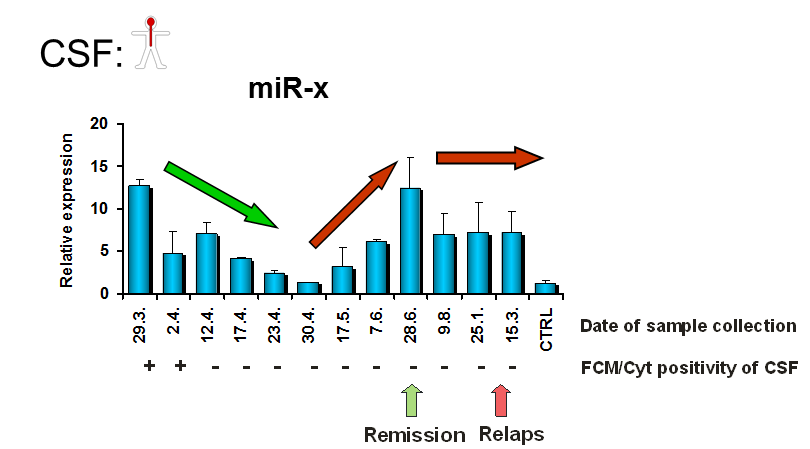
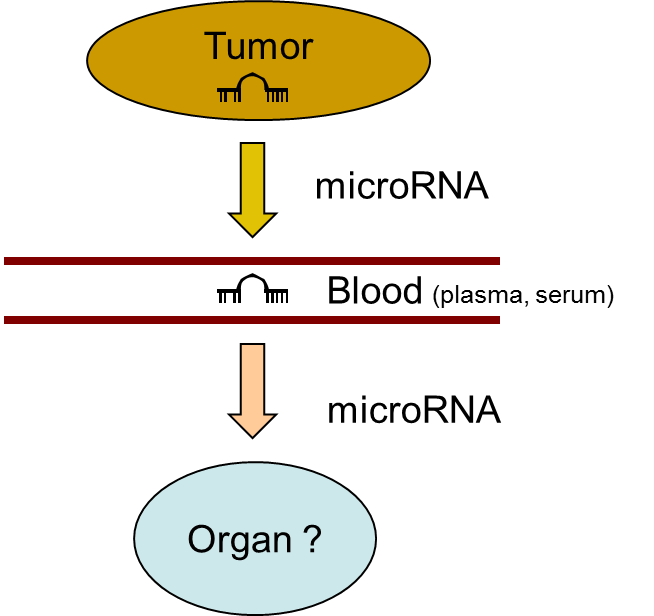
4. Role of tumor released extracellular oncogenic microRNAs in tumor biogenesis and dissemination
It is well documented that tumor-associated
microRNAs are released from the tumor into the circulation, where they
exhibit extraordinary stability. However, the biological implication of
these circulating microRNAs is unknown. Any knowledge is lacking whether
these microRNAs can enter the organs and tissues, and whether they can
aide in tumor progression and/or dissemination.
In vivo and in vitro assays answering these questions are being developed.
Methods and approaches: molecular cloning and mutagenesis, gene reporter
assays, RT-qPCR, WB, FACS, cell culturing, differentiation and transfections.
si/shRNA, mikroRNA, lncRNA. In silico anaylsis of DNA regulatory sequences
(Jaspar, Transfac, UCSC, GEO, Encode, Cage - Fantom etc). Chromatin
immunoprecipitation seq, tools for 3D interaction of chromatin – chromosome
confirmation capture (3C, Hi-C). RNA sequencing. Microscopy. Animal work –
lymphoma xenotransplantation. Analysis of leukemia patients samples.
Publications:
- Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. Sochor M, Basova P, Pesta M, Dusilkova N, Bartos J, Burda P, Pospisil V, Stopka T. BMC Cancer. 2014 Jun 18;14:448.
IF 7.276
- Aggressive acute myeloid leukemia in PU.1/p53 double-mutant mice.
Basova P1, Pospisil V, Savvulidi F, Burda P, Vargova K, Stanek L,
Dluhosova M, Kuzmova E, Jonasova A, Steidl U, Laslo P, Stopka T.
Oncogene. 2013 Oct 14. 1-11.
IF 8.559
- 5-Azacitidine in
aggressive myelodysplastic syndromes regulates chromatin structure at
PU.1 gene and cell differentiation capacity. Curik N, Burda P,
Vargova K, Pospisil V, Belickova M, Vlckova P, Savvulidi F, Necas E,
Hajkova H, Haskovec C, Cermak J, Krivjanska M, Trneny M, Laslo P,
Jonasova A, Stopka T. Leukemia. 2012 Aug;26(8):1804-11.
IF 8.966
- Epigenetic silencing of the oncogenic miR-17-92 cluster during PU.1-directed macrophage differentiation.
Pospisil V, Vargova K, Kokavec J, Rybarova J, Savvulidi F, Jonasova A,
Necas E, Zavadil J, Laslo P, Stopka T., EMBO J. 2011 Sep 6; 30,
4450-4464.
IF 10.124
- MYB transcriptionally regulates the miR-155 host gene in chronic lymphocytic leukemia.
Vargova K, Curik N, Burda P, Basova P, Kulvait V, Pospisil V, Savvulidi
F, Kokavec J, Necas E, Berkova A, Obrtlikova P, Karban J, Mraz M,
Pospisilova S, Mayer J, Trneny M, Zavadil J, Stopka T. Blood. 2011 Apr
7;117(14):3816-25.
IF 10.558
- Cdc25A localisation and shuttling: characterisation of sequences mediating nuclear export and import. Källström H, Lindqvist A, Pospisil V, Lundgren A, Rosenthal CK. Exp Cell Res. 2005. 303(1):89-100. Källström H, Lindqvist A, Pospisil V, Lundgren A, Rosenthal CK. Exp Cell Res. 2005,303(1):89-100. IF 4.148
Conferences:
- 2007 American Society of Hematology Annual meeting, Orlando, USA, oral presentation, 1st author
- 2008 American Society of Hematology Annual meeting, Atlanta, USA, oral presentation, 1st author
- 2010 American Society of Hematology Annual meeting, San Francisco, USA, oral presentation, 1st author
- 2011 International Society for Experimental Hematology Annual meeting, Vancouver, Canada, oral presentation, 1st author
- 2012 American Society of Hematology Annual meeting, San Diego, USA, 1st author
- 2013 American Society of Hematology Annual meeting, New Orleans, USA, 1st author
NEWS
- October 19-21, 2014 Cell Symposia: Regulatory RNAs, Berkley, USA
- October 28 - November 2, 2014 Keystone Symposia Conference, Cell Death Signaling in Cancer and the Immune System, Sao Paulo, Brazil
Pregraduate and PhD students are accepted and welcome.
Vítek: outdoorsports (mountaineering, skialpinism, skiing, orienteering), photography, theatre
Pavle: philosophy, swimming
Jan: curling, logical games, marbles
 Laboratory
Laboratory

Pathophysiology